

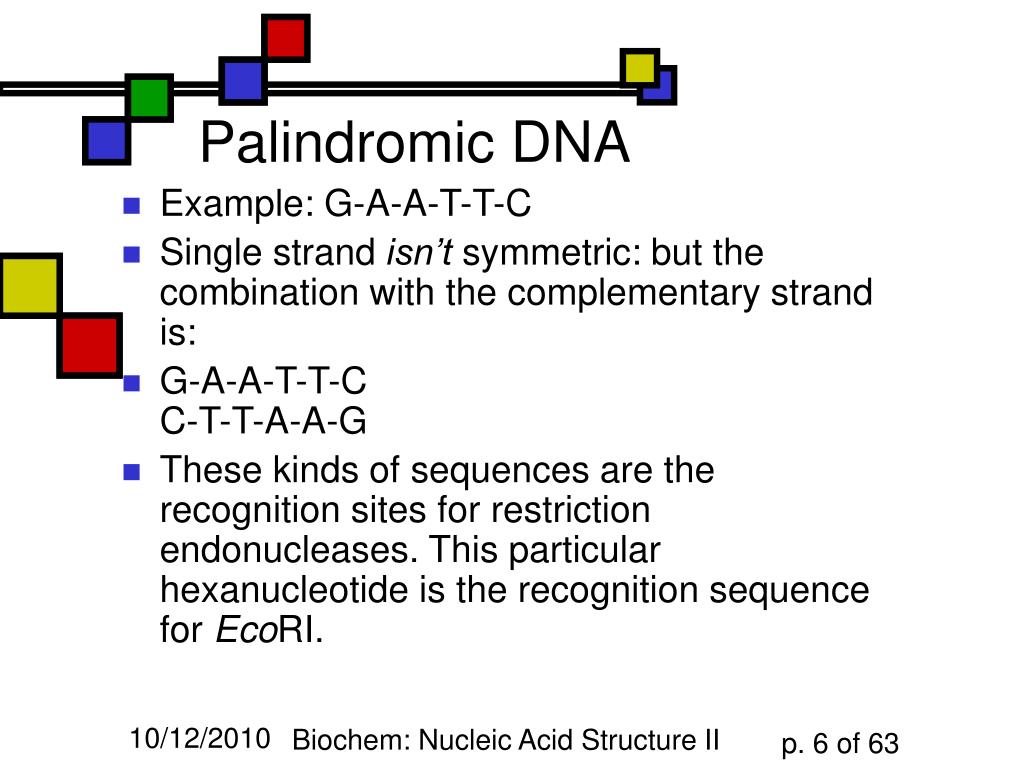
Out of the palindromic sequences of the CRISPR region RNA molecules are transcribed, which adopt a very stable arrangement (secondary structure). These sequences can be four, six or eight base pairs long, although some cutting proteins require 20 or more base pairs. DNA-cutting proteins frequently use palindrome sequences as recognition sequences, at which they cut the DNA molecule. Nevertheless, they are not entirely meaningless. Unlike word palindromes like ‘civic’ and ‘tenet’, which have a meaning, the palindromes in the genetics dictionary do not make sense and cannot be translated into functional proteins. This is the property that gives CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) its tongue-twisting name. In these sequences, the letters of the genetic code, the four base molecules adenine, cytosine, thymine and guanine, are ordered such that they have the same order as the second complementary DNA-strand – in this case read in opposite direction. The beginning of the CRISPR revolution was marked by the discovery of a large number of repeated palindromic sequences in a region of bacterial DNA. It’s worth noting that we store the answer inside the array so that we can reuse it if we reach the same state later.“Able was I ere I saw Elba”: this rather bombastic statement is a palindrome sentence, in other words it reads exactly the same forwards as it does backwards. The final answer is the maximum value between the two options. We can either move one step forward or move one step backward. If the L th and R th elements are not equal, we have two options. After that, we add 2 to the current state’s answer because we included the L th and R th elements in the palindromic subsequence. Otherwise, if both sides of the range hold equal elements, we calculate the best answer we can get from the subproblem. Also, if we’ve calculated this state before, we return its answer. We must also pass the sequence, equal to one, and equal to, where is the length of the sequence.Īs we can see, If we’ve reached a base subproblem, we return its answer. In the initial call, we must fill the array with -1 values, indicating that we haven’t calculated any state yet. Take a look at the pseudocode of the described approach. If so, we just return the calculated answer. After that, we calculate the answer to the current problem based on the answers of the subproblems.īefore dividing the problem into subproblems, we check to see if the current state has been calculated before. In each step, we recursively apply the algorithm to all the subproblems. The top-down approach is usually easier to understand. For example, from the state, if the first and n th elements are not equal, we can find the best answer in one of the ranges or. We can find the best answer by removing either the first element or the last one. The last reason is the case of the first and last elements being not equal. Therefore, from the state we can move to state if the first and the n th elements are equal. If we removed these elements, the remaining elements must form a palindromic sequence as well. The second reason is that the palindromic sequence has the first and last elements equal. Otherwise, if we reach an empty range, the answer to it is just zero. That’s because any sequence of length one is a palindrome. When we need to calculate the answer to a range of length 1, its answer equals to one. The first reason is that the base subproblem is one of two options. There are three reasons for choosing this type of subproblem. For a subproblem, we’ll find the answer to a specific range from the original sequence.
#Palindromic sequence with pallendrome full
The full problem requires finding the answer to the full sequence. Let’s think about the subproblems of our current problem.


 0 kommentar(er)
0 kommentar(er)
